Volume 13 - Year 2025 - Pages 01-10
DOI: 10.11159/ijepr.2025.001
Analysis of Genotypic and Environmental Risk Factors for the Development of Allergic Diseases in Almaty Residents
Madina Abdullayeva1,2, Nazym Altynova1*, Saida Tokmurzina1, Tamerlan Kereyev1,2, Aigerim Kassymbekova1, Yergali Kanagat1, Dmitri Gourevitch3, Danara Artygaliyeva3, Leyla Djansugurova1,2
1Institute of Genetics and Physiology
Al-Farabi Ave., 93, Almaty, Kazakhstan
2Al-Farabi Kazakh National University
Al-Farabi Ave., 71, Almaty, Kazakhstan
3Medical Center “Allergo Clinic”
Navoi 208, (Residential Complex "Shahristan")
*naz10.79@mail.ru
Abstract - Allergic diseases represent a global health problems. According to the "Association of Allergists and Clinical Immunologists" of Kazakhstan, the number of allergy sufferers in the country increases by 10-15% every year. Microarray genotyping has been widely used in allergy diagnosis due to its high throughput capability and specificity in identifying genetic markers associated with allergic diseases. This work is aimed to study the dynamics of the incidence of allergic diseases among residents of Almaty; assess the role of environmental and genotypic risk factors; identify marker genes related to asthma and allergic rhinitis and risk assessment in case and controls among patients. The findings revealed that allergic rhinitis was the most prevalent condition (58%), followed by bronchial asthma (9%). Self-reported allergy data were often inconsistent with medical diagnoses, underscoring the importance of clinical confirmation. A family history of allergies emerged as a major risk factor, significantly outweighing the impact of smoking. Genetic analysis identified specific SNPs in the IL13 and IL12B genes that were associated with increased allergy risk. For IL13, the T allele of rs1295686, A allele of rs848, and A allele of rs20541 were identified as significant. In IL12B, the T allele of rs2853694 and CC genotype of rs2569254 were linked to higher risk.
Keywords: Allergen, Microarray genotyping, SNP, Allergic rhinitis, Asthma.
© Copyright 2025 Authors - This is an Open Access article published under the Creative Commons Attribution License terms. Unrestricted use, distribution, and reproduction in any medium are permitted, provided the original work is properly cited.
Date Received: 2024-09-12
Date Revised: 2024-12-18
Date Accepted: 2025-01-02
Date Published: 2025-01-23
1. Introduction
Allergic diseases occupy a vital place in the structure of general morbidity.
8–10% of the global population suffers from one or more allergic diseases, ranging from mild rhinitis to severe anaphylaxis or asthma [1]. Allergic rhinitis (AR) is a condition affecting the nasal mucosa that arises from inflammation caused by allergens. The development of the allergic response induces pathological changes in the epithelium of the respiratory mucosa, including increased thickness, impaired ciliary function, and breakdown of the cells lining the mucosa. Consequently, this leads to impairment in nasal clearance, an increase in mucus viscosity, a reduction in the secretion capacity of cells, and drying of the mucosa.
Its prevalence in the Kazakhstani population is increasing annually, and the ecological situation is one of the main reasons for developing allergies in people. According to the "Association of Allergists and Clinical Immunologists" of Kazakhstan, the number of allergy sufferers in the country increases by 10-15% yearly. This increases the risk of developing severe allergies that can lead to disability. Approximately one million individuals in Kazakhstan experience bronchial asthma (BA), with a higher prevalence among children.
This study offers important insights into allergic diseases and their underlying factors in Kazakhstan. It has significant implications for patient treatment and future research in this area. While genetic databases provide information on certain populations, there is little data on genetic variations specific to Kazakhs or Central Asians. Comprehensive genome-wide association studies have not been conducted in this region before, making this study a pioneering project.
2. Host/Genetic risk factors for the development of allergic rhinitis
The characteristics of an individual, including race, sex, age, and heredity influence the allergy risk. The most significant among these factors is heredity.
The advent of Genome-Wide Association Studies (GWAS), which is an unbiased approach to investigating a vast number of typical variations throughout the genome in both healthy and diseased individuals, has significantly transformed our comprehension of allergic disease genetics. Identifying susceptibility genes linked with BA and AR gives confidence that common and distinctive genetic locations are associated with them, thereby offering new perspectives into potential pathways and mechanisms underlying these diseases. About 30% of people with rhinitis also have asthma, and more than 80% of those with asthma experience symptoms of rhinitis [2]. This connection highlights how closely related these conditions are and suggests they may share some of the same genetic and biological causes.
Race: The reasons for hay fever and asthma among different races are complex, as it is challenging to distinguish between environmental influences, migratory patterns, and racial factors. Studies have found that black individuals exhibit elevated levels of IgE compared to their Caucasian counterparts [1].
Heredity: Heredity is believed to play a significant role in developing most allergic conditions. A genetic epidemiologic study conducted in 2018 in Jiangsu province, China, involving 23,825 families, found that the average heritability of allergic rhinitis (AR) was 81.86% across the first, second, and third generations [3].
Gender: The variation in gender can be clarified by the greater sensitization rate of grass pollen and house dust mites among men than women. This can also explain the higher likelihood of boys developing asthma. Although this dissimilarity tends to decrease with advancing age, most researchers assert that males exhibit more prevalent specific IgE antibodies, skin test positivity, and elevated total IgE levels compared to females [1].
Age: According to research, the highest levels of IgE are observed during infancy and decline rapidly between the ages of 10 and 30. After that, their rate of decrease becomes gradual. Asthma is more frequent in children under ten, whereas hay fever primarily affects young adults and children [1].
3. Environmental risk factors for the development of allergic diseases
The rise in allergic sensitivity and its prevalence is often attributed to environmental factors, including heightened levels of air pollution, changes in lifestyle habits, and a reduction in bacterial/viral infections. Allergic rhinitis is frequently caused by indoor allergens present in household surroundings.
Indoor allergens: Allergic diseases like asthma and allergic rhinitis are primarily triggered by house-dust mites (HDM), the most common indoor allergens. According to worldwide studies, Dermatophagoides pteronyssinus and Dermatophagoides farinae are two of the most widespread mite species responsible for these allergies [4]. The main problem lies in creating a low-allergen environment within patients' homes.
Air Pollution and outdoor allergens: Air pollution may increase the concentration of pollen allergens to which individuals may be exposed. Studies provide evidence that the amount and composition of pollen have undergone substantial changes due to climate-related factors. As reported in sources, a widely accepted phenomenon is that tree and grass pollen can become significantly more allergenic under high carbon dioxide concentrations. Pollutants irritate the exposure area and initiate complex immune mechanisms (see Figure 1) [5].
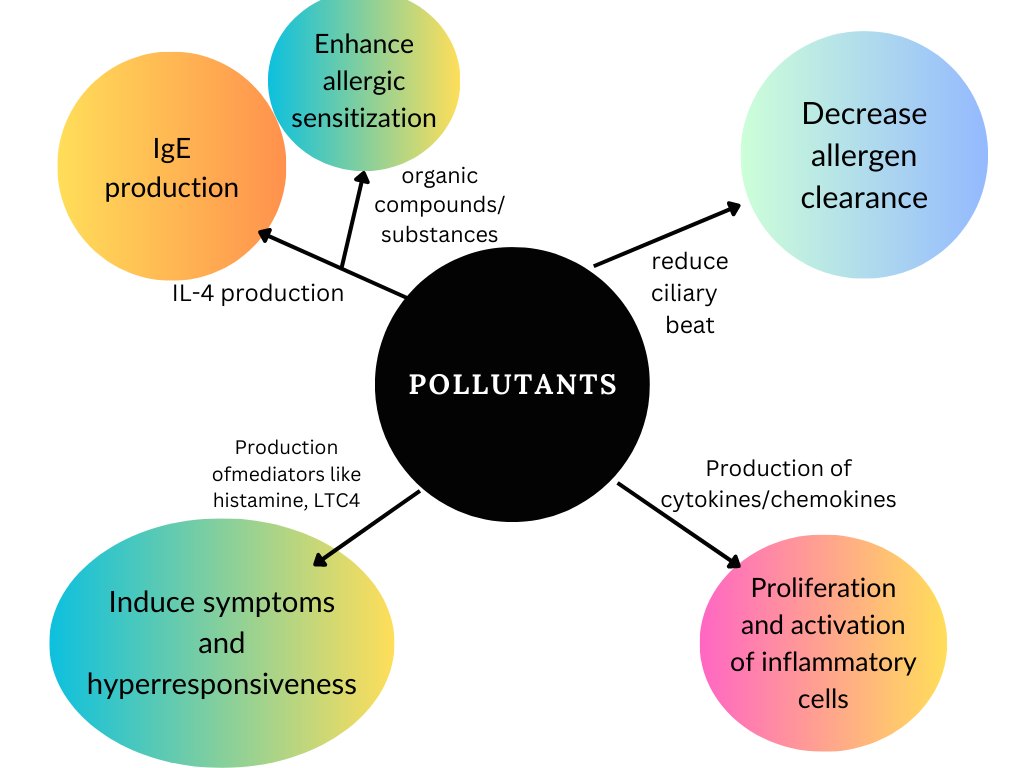
Both indoor and outdoor allergens and pollutants need to be considered. These include:
- Biologic allergens (dust mites, cockroaches, animal dander, and mold)
- Environmental tobacco smoke-smoking during pregnancy and after delivery is related to a substantial risk of developing asthma
- Irritant chemicals and fumes-traffic pollution and high ozone levels
- Products from combustion devices
Effect of smoking: The smoke from cigarettes is filled with harmful substances and potent triggers that cause inflammation. It's commonly known to be a significant contributor to respiratory and allergic diseases, making active smoking and second-hand exposure risky for adolescents who may develop asthma or allergic rhinitis [6]. Moreover, mice exposed to tobacco smoke had increased sensitivity to allergens and weaker immune responses in their airways [4].
Other factors: Having a pet can significantly increase the likelihood of developing sensitivity to them. Families with pets tend to have higher rates of asthma, rhinitis, and skin allergies than those without animals [4]. Hence, individuals already sensitized and experiencing these conditions may benefit from removing their cat or dog from their household environment.
The allergic disease has been linked to various factors such as alterations in lifestyle, dietary changes, geographical disparities, climate fluctuations, economic situations, family history, or structure. Other causes include the method of infant feeding and overexposure to allergens during infancy, and smoking cigarettes.
4. GWAS in allergy research
There is a steep upward trend in the prevalence of allergic diseases worldwide. Such allergies include allergic rhinitis, atopic dermatitis, asthma, and others. Although allergies have been studied for over a hundred years, the research faced obstacles in gene identification. However, recent advances in GWAS enabled scientists to understand the underlying genetic mechanisms of allergies [7].
In 2007 Moffatt et al. completed the first GWAS on asthmatics. The research group identified significant associations in 17q21. Since the first GWAS of asthma found variants at the 17q21 locus linked to ORMDL3 expression, this region has been the most studied and confirmed.
About half of the published GWAS studies on asthma didn't find any significant genetic associations in their discovery phase, and most of these studies had fewer than 2,000 participants. This suggests that larger sample sizes (≥10,000) are needed to identify genetic factors linked to asthma [8].
There is limited data regarding allergic rhinitis GWAS. In studies of AR, it was shown that several genes are associated with different immune-related diseases, including allergies and autoimmune disorders. These genes include SDAD1, CXCL10, CXCL9, RANTES, CXCL11, IL1R1, IL13, IL18, IL21/IL2, IL23R, IL12RB1, IL27, C11orf30, SMAD3, TLR1, GATA3, and HLA-DQ [9].
The review of allergy and asthma GWAS studies done by Portelli et al. concluded that genes that affect both the immune system and epithelial function suggest the airway epithelium plays an important role in asthma. Additionally, the differences between genetic factors linked to IgE levels and asthma show the complexity of these diseases [4].
Genetic studies on allergies reveal both shared and unique loci across ethnic groups, emphasizing the impact of ancestry on disease susceptibility. For example, variants in immune-related genes like HLA show population-specific effects, while some loci are common across diverse ancestries [10].
5 Materials and Methods
The study focused on 85 patients in Almaty who had different types of allergic diseases and sought treatment at the private clinic "Allergo Clinic." Their diagnoses were made by qualified specialists using clinical, laboratory, and research data based on established criteria. A cohort of 85 healthy individuals from Almaty and the surrounding regions was used as a control group.
Questionnaires provided by the clinic are accompanied by personal data and clinical health survey data served as materials for this study. Biomaterials were chosen from individuals with no history of cancer, heart disease, neurological conditions, autoimmune disorders, allergies, diabetes, or metabolic/thyroid dysfunction. DNA storage occurs in single-use test tubes coated with EDTA. EDTA ensures the storage of genetic material.
The ReliaPrep™ Blood gDNA Miniprep System (Promega, USA) was used for this study, ensuring high-purity DNA through a streamlined process of lysis, binding, washing, and elution. Blood samples were lysed at 65°C for 10 minutes, releasing DNA, which was then bound to a silica membrane via centrifugation. After washing impurities, purified DNA was eluted and assessed using a NanoDrop spectrophotometer, ensuring samples met genotyping quality standards (>50 mg/ng).
Genotyping was conducted using the Infinium HTS Assay (Illumina, USA), capable of analyzing up to 750,000 SNPs per sample. Genomic DNA underwent amplification, enzymatic fragmentation, and precipitation before being resuspended for hybridization. The Infinium ImmunoArray-24 v2 BeadChip, designed to detect immune-related genetic variants, was used for hybridization, where complementary DNA strands bound to probes on the BeadChip.
Hybridized DNA underwent extension and staining with fluorescent dyes, enabling precise SNP detection. The iScan System was employed for high-throughput imaging, scanning up to 96 samples per batch. This comprehensive workflow ensured reliable genotyping for downstream analysis, contributing to the identification of genetic variants linked to allergic diseases.
5.1 Data Processing, Visualization, and Analysis
Survey data was organized into Excel sheets and visualized using R, enabling compact and interpretable presentations for further analysis.
Genotype data from iScan was analyzed using the GenomeStudio Genotyping Module. Samples with <98% call rates were excluded after quality control. PLINK, a robust genome-wide association toolset, processed genotypes efficiently, identifying variants across large datasets.
Annotated variants were cross-referenced with key databases:
dbSNP: Provided genomic location, gene impact, and functional effects.
ClinVar: Identified associations with known diseases.
1000 Genomes Project: Evaluated population-specific variant frequencies.
5.2 Statistical Analysis
Case-control cohorts were evaluated for demographic and behavioral consistency using traditional statistical methods, with significant differences identified at p<0.05 via chi-square tests.
Odds Ratio (OR) Calculation
ORs and 95% confidence intervals (CIs) were calculated to determine the relative risk of mutant versus normal genotypes between allergy and control groups using standard formulas:

Where:
a - the number of people in the allergy group with the mutant genotype;
b - the number of people in the allergy group who have a normal genotype;
c - the number of people in the control group with the mutant genotype;
d - the number of people in the control group with a normal genotype.
The formulas calculated the reliable confidence interval:
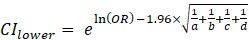
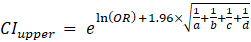
Where 1.96 is a Z-value for a 95% confidence level.
Results and Discussion
6.1 Characteristics of the study and control groups
Allergic diseases are multifaceted and intricate, resulting from genetic predisposition and environmental factors. Scientists worldwide are engaged in exploring genes accountable for the formation of allergies. A genome-wide association analysis was made for the first time in an attempt to analyze allergic disorders in Almaty's population comprising Kazakhs (75.29%), Russians (12.94%), and other ethnic groups (5.88%) (see Table 1).
Blood samples were taken from 85 patients with AR and BA to identify candidate genes for allergy predisposition. The mean age at enrolment for case group - 30.8 years, control group - 44.7 years.
Table 1. Characteristics of control and case subjects
|
Characteristics |
Case |
Control |
p-value |
|
|
n = 85 (%) |
n=85 (%) |
|
|
Onset age |
|||
|
0–20 |
16 (18.8) |
0 (0) |
|
|
21-40 |
52 (61.2) |
62 (72.9) |
|
|
41-60 |
15 (17.6) |
19 (22.3) |
|
|
61+ |
2 (2.3) |
4 (4.7) |
0.8370 |
|
Gender |
|||
|
Male |
30 (35.3) |
29 (34.1) |
|
|
Female |
55 (64.7) |
56 (65.9) |
0.8720 |
|
Nationality |
|||
|
Kazakhs |
64 (75.3) |
77 (90.6) |
|
|
Russians |
11 (12.9) |
5 (5.9) |
|
|
Others |
10 (11.7) |
3 (3.5) |
0.0271* |
|
Current smokers |
|||
|
Yes |
12 (14.1) |
8 (9.4) |
|
|
No |
73 (85.9) |
77 (90.6) |
0.2438 |
|
Family history of allergic rhinitis |
|||
|
Yes |
50 (58.8) |
25 (29.4) |
|
|
No |
35 (41.2) |
60 (70.6) |
0.0001* |
*Significant at p < 0.05
The control group was selected based on previous studies from the genetic bank of the Laboratory of Molecular Genetics at the Institute of Genetics and Physiology. This group comprised healthy individuals matched by gender, ethnicity, and age. Analysis revealed no history in this group related to oncological, cardiovascular, neurological, or autoimmune conditions or allergies.
Based on the results, it can be concluded that the groups of individuals with allergic diseases and those without are suitable for this molecular epidemiological study using a case-control approach. The patients were categorized based on clinical examinations of allergic diseases.
6.2 Prevalence and characteristics of allergic diseases
According to a self-report of the estimated 85 subjects, the prevalence rate of allergic diseases overall was 85.88% (73 people), and the rate of allergic rhinitis and bronchial asthma were 37.65%, and 14.12%, respectively.
As for the co-occurrence of two or more allergic diseases, rhinitis/dermatitis and rhinitis/bronchial asthma accounted for 2.3% (2/85) and 7.06% (6/85), respectively. As for the co-occurrence of three allergic diseases, AR/BA/hives accounted for 3.53% (3/85). The least prevalent was for bronchial asthma/hives, 1.18% (1/85) of all subjects. 14.12% of participants (12/85) did not answer.
The self-report of symptoms could not be judged directly; hence the doctor's diagnosis would serve as an approval. For example, the number of patients diagnosed with rhinitis (58%) is higher than the self-reported cases (38%). As for those patients who were not diagnosed (or whose diagnosis was under question), genotyping could help to detect disease (see Figure 2).

As the results showed, self-report cannot show the actual incidences of allergic diseases. The best verification for self-reported allergic diseases was achieved by combining the information on self-reported diseases and the medical records. After the verification, it was revealed that the most prevalent form of allergic disease is allergic rhinitis among the patients. It is for patients with two allergic diseases - allergic rhinitis and hives. No patient with three allergic diseases was identified in our study.
6.3 Identification of potential factors
6.3.1 Smoking
The effects of smoking and positive family history on the development of allergic rhinitis were determined. In Figure 3, it is shown that smokers (current) versus never smokers were not significantly related to the risk of allergic diseases in the univariate analyses (OR 1.16; 95% CI, 0.50-2.70). The results indicate a weak and statistically non-significant association between smoking and allergic diseases (OR: 1.16, 95% CI: 0.50–2.70).
There is also a study done by Hisinger-Mölkänen H. et al., current smoking, without exposure to occupational irritants, yielded an OR of 1.22 (95% CI 1.00–1.49) for allergic rhinitis, showing that there is a weak association [11].
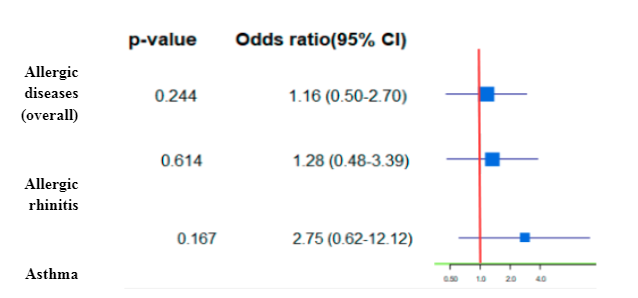
Therefore, while the study provides some evidence for a potential link between smoking and allergic diseases, the findings are inconclusive and need to be interpreted with caution. Further research with larger sample sizes and more rigorous study designs is needed to confirm the association and establish the direction of causality between smoking and allergic diseases.
6.3.2 Positive family history
Positive family records of allergic diseases gave the high-risk estimates: OR 3.43 (95% CI 1.81-6.47) for allergic diseases, 5.28 (95% CI 2.45 - 11.38) for allergic rhinitis, and 7.20 (95% CI 1.36–38.13) for bronchial asthma (see Figure 4).
The study found that children with a family history of allergies are more likely to develop allergic diseases such as rhinitis and asthma. The odds ratios show that these individuals have 3.43, 5.28-, and 7.20-times higher likelihood than those without a family history of allergy for developing the respective conditions mentioned earlier. It is crucial to take into account the patient's family medical background when diagnosing or treating an allergic disease based on these outcomes.
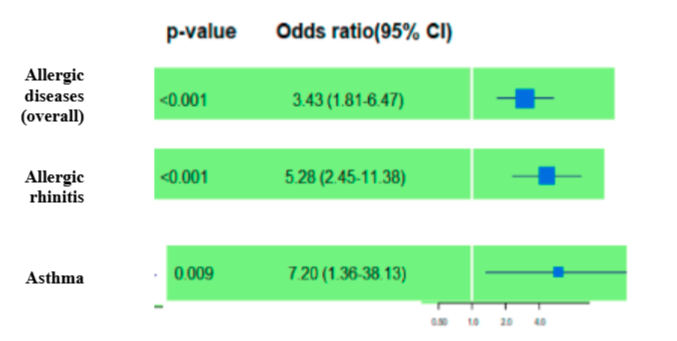
Our results correlate with the study done in China by Min Y. et al., where family history of allergic diseases (2.23, 1.92 to 2.60, 1.66E-10) were found to be independently and consistently associated with the significant risk of childhood allergic diseases overall and by clinical manifestations. Without adjustment asthma yielded OR 2.47(2.14, 2.85) and rhinitis yielded OR 3.30(3.02, 3.62) [12]. Also, the study done by Alatawi et al. in Saudi Arabia showed that a family history of asthma or atopic conditions 3.118 times increased the likelihood of allergic conditions (OR = 3.118, 95% CI: 1.827-5.320) [13].
This study found that smoking and a family history of allergic diseases are potential factors in predicting the risk of allergic diseases. Family history was a significant factor in improving risk prediction capability, especially for clinical manifestations.
6.4 Performing GWAS
This type of research is called a Genome-wide Association Study (GWAS), a promising tool for evaluating SNP associations to various diseases and complex traits. The study carried out a genome-wide microarray genotyping that focused on identifying SNPs on the iScan platform via The Infinium ImmunoArray-24 v2. Eighty-five control samples and eighty-five allergy case samples were genotyped. The protocol required the exclusion of samples with call rates <98%. Thus, two allergy samples were excluded, while the other 83 DNA samples passed the call rate requirements. GenomeStudio was the primary analysis tool that determined quality and corrected clusterization errors.
According to the genotyping results, the Infinium ImmunoArray identified 17 genes and 43 single nucleotide polymorphisms related to allergies, including allergic rhinitis and asthma. Some details are worth noting in the said polymorphisms: 8 SNPs show missense mutations, while 34 were silent, and one indicated a synonymous mutation. Regarding the exome, the genotyping identified 14 exonic polymorphisms that showed both silent and missense mutations. The following review includes risk associations of four genes.
6.5 IL13 gene
Activated T-helper 2 (Th2) cells cause allergic inflammation and the development of atopic asthma. Interleukin 13 (IL13) is a cytokine that participates in cell signaling and immune response regulation primarily produced by Th2. Our results show three significant IL13 polymorphisms (rs1295686, rs20541, rs848) linked to allergic disorders such as asthma and allergic rhinitis.
As shown in Figure 5, the SNPs significantly correlate with asthma risk in this case study. The results signify the role of alleles in rs1295686 (allele T– OR=3.34, 95% CI=2.13-5.22, p=<0.001), rs20541 (allele A - OR=1.39, 95% CI=1.03-1.86, p=0.03). The odds of having asthma (rs1295686) and either or both asthma and allergic rhinitis (rs20541) are substantially higher among individuals with the risk alleles than healthy individuals, which the multiplicative model of inheritance supports.
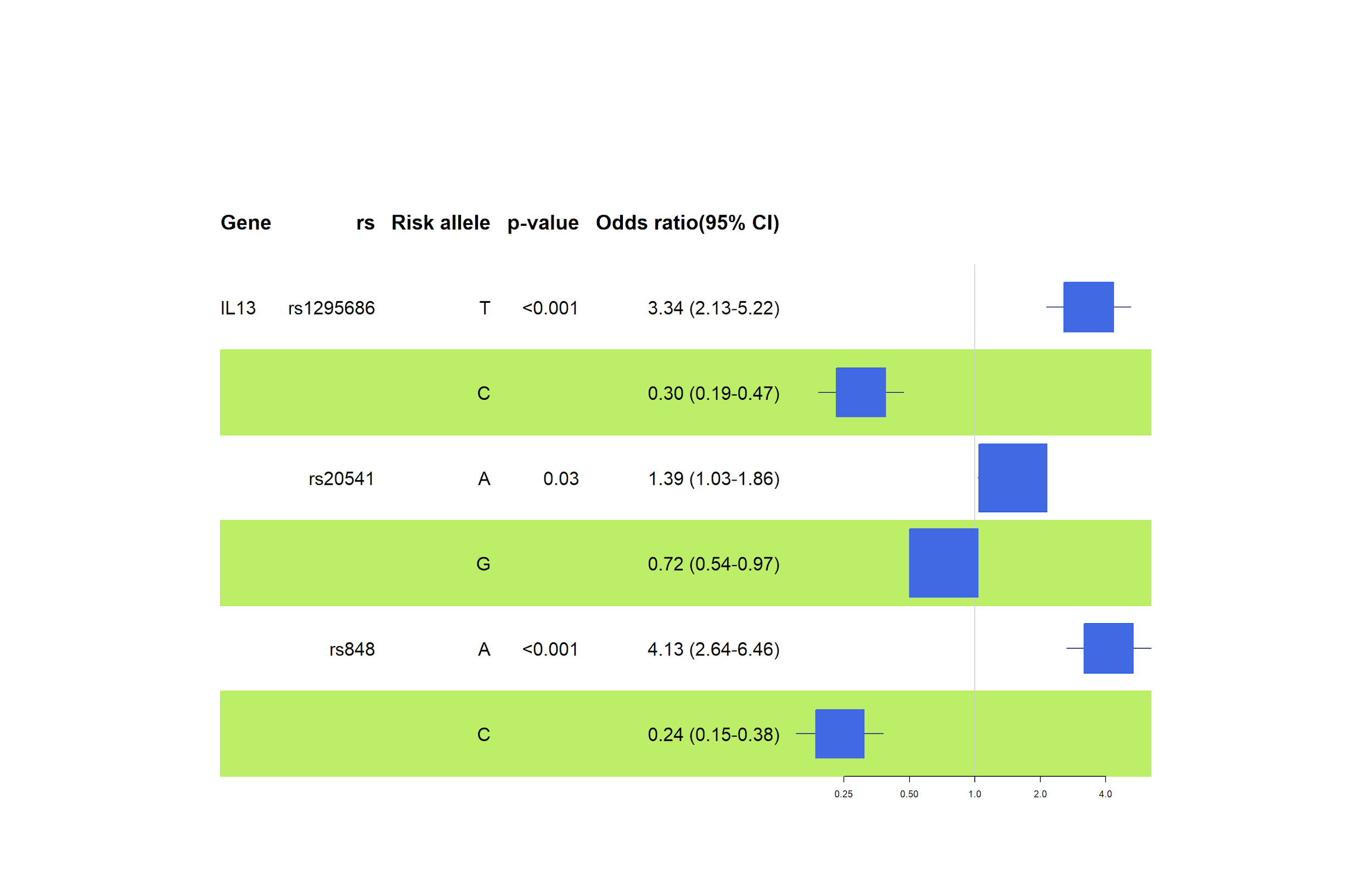
Furthermore, this genetic association study confirmed the link of the rs848 polymorphism with atopic asthma and allergic rhinitis (hay fever). The results suggest that rs848 (OR=4,13, 95% CI= 2.64-6.46, p=<0.001) poses a significant risk in individuals with the risk allele (A), as shown in Figure 6. According to Halwani et al., similar results were obtained concerning the Saudi Arabian population, where risk alleles for rs1295686 and rs20541 confirmed higher odds of developing asthma in the presence of either allele [14].
As illustrated in Figure 6, the association analysis of individual genotypes, according to the general inheritance model, revealed increased risks caused by homozygotes in rs1295686 (TT genotype - OR=5.28, 95% CI=2.50-11.14, p=<0.001) and rs848 (AA genotype - OR=4.70, 95% CI=2.30- 9.61, p=<0.001). Our results for rs2451 showed somewhat elevated risks in GA and AA genotypes, but the statistic was lost in the confidence intervals.
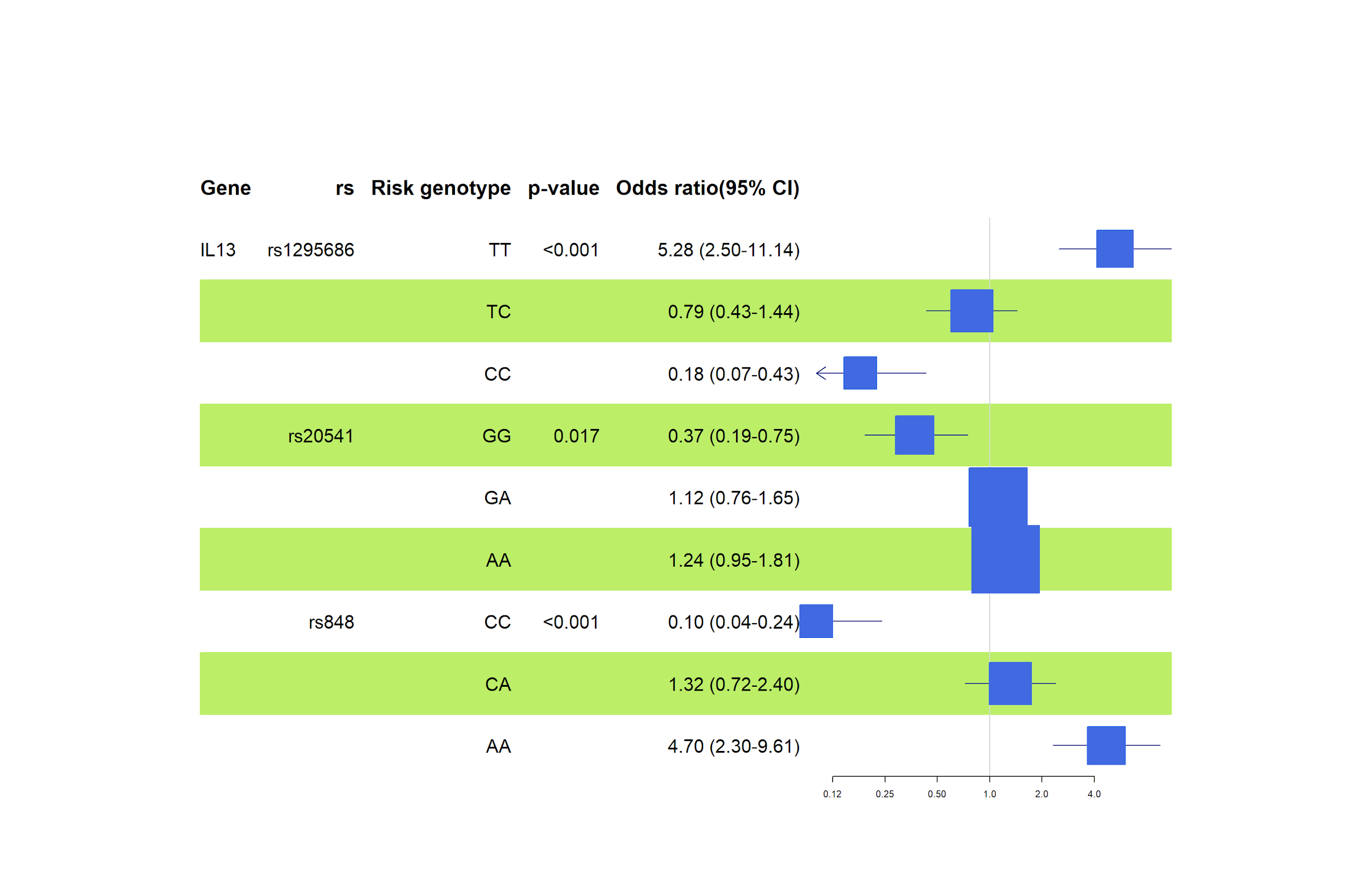
However, a similar study (Halwani et al.) found that the risk genotype frequencies among Saudi Arabian asthmatics and controls indicate a significant risk for rs20451 and not rs1295686 [14]. Meanwhile, Halwani's study ignored rs848. Resende et al. reported less risk for asthma susceptibility in rs20541 (GG genotype) among the Portuguese population [15].
3.6 IL12B gene
Analysing the rs3212227 variation (TT genotype - OR=1.60, 95% CI=0.87 – 2.94, p=0.299), we see a high odds ratio, but the statistic was lost in the confidence interval and p-value. We observed that asthma or allergic rhinitis onset risk is not statistically significant for either genotype in rs3212227, as shown in Figure 7. There is no significant effect between the variables under this investigation, possibly due to the small scale of this investigation.
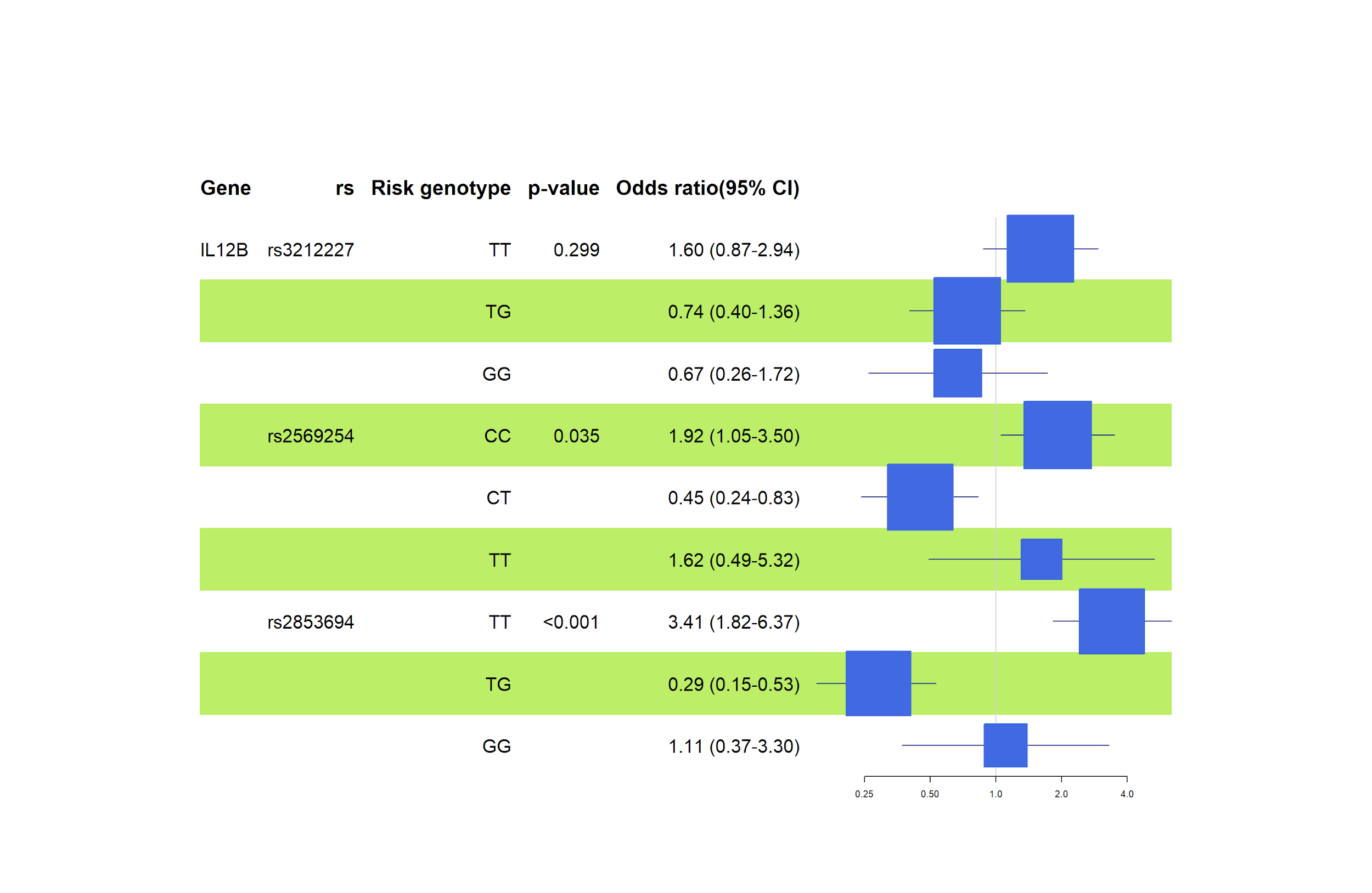
Studies show no strong association between the IL-12B rs3212227 polymorphism and either, asthma or allergic rhinitis. For example, research in Taiwan found no difference in the frequency of this gene variant between people with asthma and those without, although another gene, IL-12A rs568408, was linked to more severe asthma symptoms [16]. Similarly, a study on allergic rhinitis in Iran found no association between IL-12B rs3212227 and the condition, suggesting this specific genetic variant might not significantly influence allergy risks [17].
According to the general inheritance model, rs2569254 (CC genotype - OR=1.92, 95% CI =1.05 – 3.50, p=0.035) showed a high-risk susceptibility to allergic rhinitis in our test subjects (see Figure 7). Despite a small pool of samples, the results are compelling and statistically significant for the rs2569254 association. Zhang et al. reported similar results that showed the single effect of rs2569254 in Chinese individuals with allergic rhinitis while confirming the combined effect of rs2569254 in IL12B, rs8193036 in IL17A gene, and rs1898413 in RORα on allergic rhinitis [18].
Additionally, the results suggest that rs2853694 (TT genotype - OR=3.41, 95% CI=1.82 – 6.37, p=<0.001) is a risk polymorphism for asthma (Figure 7). According to Figure 12, SNP rs2853694 (OR=1.97, 95% CI =1.24 – 3.13, p=0.004) showed a significant association with asthma in individuals with risk (T) alleles under this investigation. The odds ratios confirm a significant risk of the T allele at this locus of developing asthma. On the other hand, Accordini et al. analyzed 326 asthma cases in Verona, and their results suggested a non-significant moderate association among their samples regarding symptom severity [19].
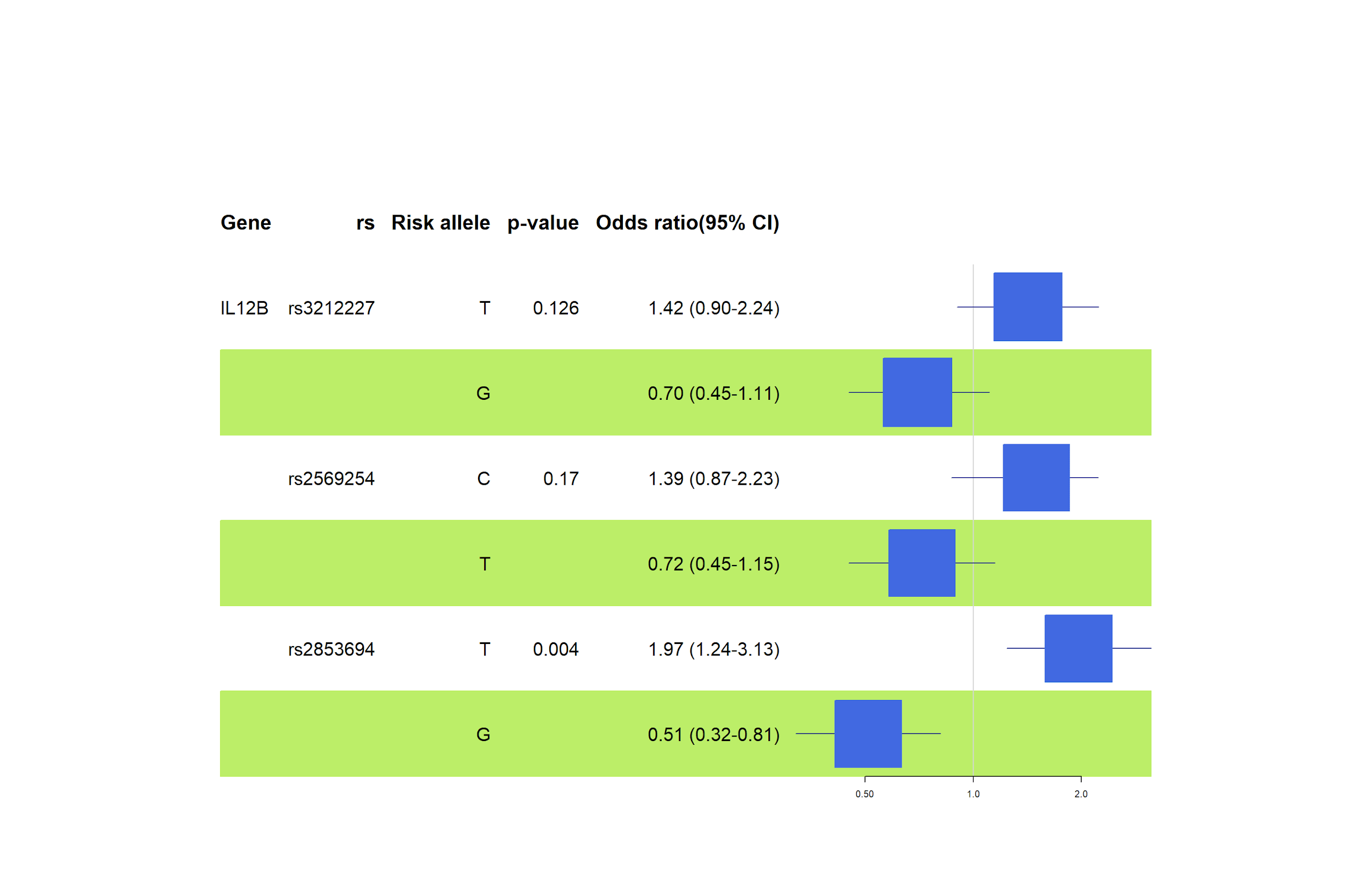
Moreover, rs3212227 did not show any statistically significant association with asthma, whereas rs2569254 (C allele - OR=1.39, 95% CI =0.87 – 2.23, p=0.17) scored a higher odds ratio but lost the statistic in confidence interval and p-value (see Figure 8). IL12B rs2569254 polymorphisms require more investigation.
3.8 Limitations
A frequent problem while studying multifactorial diseases is the discrepancies in findings between various studies. Ancestry backgrounds, homogenous samples, small sample size, epigenetics, and environments may cause the result differences.
Our data showed some irregularities with statistics. The limited number of relevant studies in this area made it challenging to directly compare and support our findings. For future reference, the sample size must be expanded.
5. Conclusion
This study aimed to assess the prevalence of allergic diseases based on survey data and to identify potential genetic and environmental risk factors contributing to these conditions. The findings reveal that allergic rhinitis, with a prevalence rate of 58%, is the most common allergic disease, followed by bronchial asthma at 9%, based on medical diagnoses. Notably, self-reported data from patients did not accurately reflect the true incidence of allergic diseases, underscoring the importance of clinical verification.
A forest plot analysis highlighted that a positive family history of allergic diseases (OR 3.43, 95% CI 1.81–6.47) is a more significant risk factor than smoking (OR 1.16, 95% CI 0.50–2.70) for developing allergies. A comprehensive review of six genes involving nine single nucleotide polymorphisms (SNPs) revealed that five polymorphisms showed significant associations with asthma and allergic rhinitis in either genotypic or allelic forms.
In the IL13 gene, rs1295686 (T allele: OR 3.34, TT genotype: OR 5.28), rs848 (A allele: OR 4.13, AA genotype: OR 4.70), and rs20541 (A allele: OR 1.39) were linked to allergies. Similarly, in the IL12B gene, rs2853694 (T allele: OR 1.97, TT genotype: OR 3.41) and rs2569254 (CC genotype: OR 1.92) suggested high-risk associations with allergic diseases.
A complete genome-scale bioinformatic analysis will continue in frame of the project “Analysis of environmental risks and genetic predisposition to the development of bronchial asthma among residents of Almaty and the Almaty region" (AP23488865), funded by the Ministry of Science and High Education of the Republic of Kazakhstan.
References
[1] F. Aldakheel, "Allergic Diseases: A Comprehensive Review on Risk Factors, Immunological Mechanisms, Link with COVID-19, Potential Treatments, and Role of Allergen Bioinformatics," International Journal of Environmental Research and Public Health, vol. 18, no. 22, p. 12105, Nov. 2021. View Article
[2] E. Nappi, G. Paoletti, L. Malvezzi, S. Ferri, F. Racca, M. R. Messina, F. Puggioni, E. Heffler, and G. W. Canonica, "Comorbid allergic rhinitis and asthma: important clinical considerations," Expert Rev. Clin. Immunol., vol. 18, pp. 747-758, 2022. View Article
[3] L. Cheng, J. Chen, Q. Fu, S. He, H. Li, Z. Liu, G. Tan, Z. Tao, D. Wang, W. Wen, R. Xu, Y. Xu, Q. Yang, C. Zhang, G. Zhang, R. Zhang, Y. Zhang, B. Zhou, D. Zhu, L. Chen, X. Cui, Y. Deng, Z. Guo, Z. Huang, Z. Huang, H. Li, J. Li, W. Li, Y. Li, L. Xi, H. Lou, M. Lu, Y. Ouyang, W. Shi, X. Tao, H. Tian, C. Wang, M. Wang, N. Wang, X. Wang, H. Xie, S. Yu, R. Zhao, M. Zheng, H. Zhou, L. Zhu, and L. Zhang, "Chinese Society of Allergy Guidelines for Diagnosis and Treatment of Allergic Rhinitis," Allergy Asthma Immunol. Res., vol. 10, no. 4, pp. 300-353, 2018. View Article
[4] M. Portelli, E. Hodge, and I. Sayers, "Genetic risk factors for the development of allergic disease identified by genome wide association," Clinical & Experimental Allergy, vol. 45, 2014. View Article
[5] T. Mørkve Knudsen, F. Rezwan, Y. Jiang, W. Karmaus, C. Svanes, and J. Holloway, "Transgenerational and intergenerational epigenetic inheritance in allergic diseases," Journal of Allergy and Clinical Immunology, vol. 142, no. 3, pp. 765-772, Sep. 2018. View Article
[6] North ML, Jones MJ, MacIsaac JL, Morin AM, Steacy LM, Gregor A, Kobor MS, Ellis AK., "Blood and nasal epigenetics correlate with allergic rhinitis symptom development in the environmental exposure unit," Allergy, Jan 2018. View Article
[7] M. A. Portelli, E. Hodge, and I. Sayers, "Genetic risk factors for the development of allergic disease identified by genome‐wide association," Clinical & Experimental Allergy, vol. 45, no. 1, pp. 21-31, 2015. View Article
[8] K. W. Kim and C. Ober, "Lessons Learned From GWAS of Asthma," Allergy Asthma Immunol. Res., vol. 11, no. 2, pp. 170-187, Mar. 2019. View Article
[9] B. Y. Choi, M. Han, J. W. Kwak, and T. H. Kim, "Genetics and Epigenetics in Allergic Rhinitis," Genes, vol. 12, no. 12, Art. no. 2004, 2021. View Article
[10] Shirai, Y., Nakanishi, Y., Suzuki, A., Konaka, H., Nishikawa, R., Sonehara, K., Namba, S., Tanaka, H., Masuda, T., Yaga, M., Satoh, S., Izumi, M., Mizuno, Y., Jo, T., Maeda, Y., Nii, T., Oguro-Igashira, E., Morisaki, T., Kamatani, Y., Nakayamada, S., Nishigori, C., Tanaka, Y., Takeda, Y., Yamamoto, K., Kumanogoh, A., Okada, Y., "Multi-trait and cross-population genome-wide association studies across autoimmune and allergic diseases identify shared and distinct genetic components," Annals of the Rheumatic Diseases, vol. 81, pp. 1301-1312, 2022. View Article
[11] H. Hisinger-Mölkänen, P. Piirilä, T. Haahtela, A. Sovijärvi, and P. Pallasaho, "Smoking, environmental tobacco smoke and occupational irritants increase the risk of chronic rhinitis," World Allergy Organ J., vol. 11, no. 6, 2018. View Article
[12] M. Yang, X. Deng, S. Wang, K. Wang, W. Niu, and Z. Zhang, "Risk factors for allergic diseases: a cross-sectional survey of 9,501 Chinese preschool-aged children," Transl Pediatr, vol. 10, no. 8, 2021. View Article
[13] A. M. Alatawi, A. M. Alanazi, A. B. S. Almutairi, R. F. A. Albalawi, A. A. M. Alhakami, A. A. S. Alnuaman, L. D. D. Alzahrani, Z. S. Albalwi, A. A. H. Alabawy, L. M. M. Aljohani, and N. A. S. Alatawi, "Prevalence and Risk Factors of Allergic Diseases Among School Students in Tabuk: A Cross-Sectional Study," Cureus, vol. 15, no. 3, p. e36658, Mar. 2023. View Article
[14] R. Halwani, A. Vazquez-Tello, R. Kenana, M. Al-Otaibi, K. A. Alhasan, Z. Shakoor, S. Al-Muhsen, "Association of IL-13 rs20541 and rs1295686 variants with symptomatic asthma in a Saudi Arabian population," Journal of Asthma, vol. 55, no. 11, pp. 1157-1165, Nov. 2018. View Article
[15] E. P. Resende, A. Todo-Bom, C. Loureiro, A. M. Pinto, B. Oliveiros, L. Mesquita, and H. C. Silva, "Asthma and rhinitis have different genetic profiles for IL13, IL17A and GSTP1 polymorphisms," Rev. Port. Pneumol. (Engl. Ed.), vol. 23, no. 1, pp. 10-16, 2017. View Article
[16] C. T. Shen, C. W. Tsai, W. S. Chang, S. Wang, C. Y. Chao, C. L. Hsiao, W. C. Chen, T. C. Hsia, and D. T. Bau, "Association of Interleukin-12A rs568408 with Susceptibility to Asthma in Taiwan," Scientific Reports, vol. 7, no. 1, p. 3199, 2017. View Article
[17] S. Falahi, F. Salari, A. Rezaiemanesh, S.H. Mortazavi, F. Koohyanizadeh, R. Lotfi, A.G. Karaji, "Association of interleukin-12B rs6887695 with susceptibility to allergic rhinitis," Immunologic Research, vol. 69, no. 2, pp. 189-195, Apr. 2021 View Article
[18] Y. Zhang, J. Li, C. Wang, and L. Zhang, "Association between the interaction of key genes involved in effector T-cell pathways and susceptibility to develop allergic rhinitis: A population-based case-control association study," PLoS One, vol. 10, no. 7, p. e0131248, Jul. 2015. View Article
[19] S. Accordini, L. Calciano, C. Bombieri, G. Malerba, F. Belpinati, A. R. Lo Presti, A. R. Baldan, M. Ferrari, L. Perbellini, R. de Marco, "An Interleukin 13 Polymorphism Is Associated with Symptom Severity in Adult Subjects with Ever Asthma," PLoS One, vol. 11, no. 3, p. e0151292, Mar. 2016. View Article

